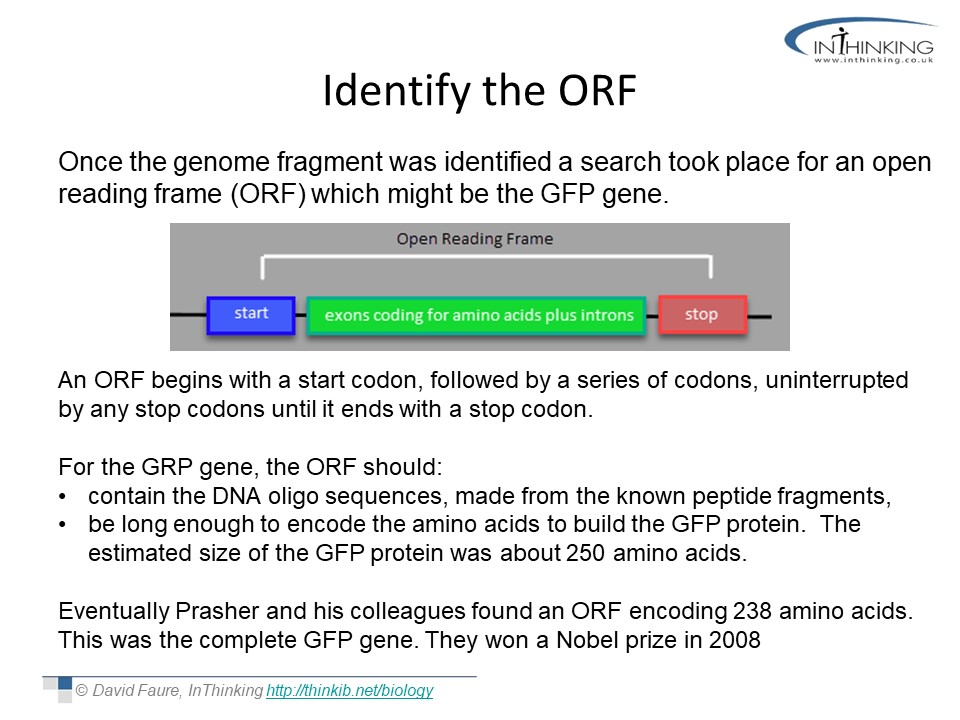
ORF activity
 The concept of Open Reading Frames seems quite complex at first, but this activity with it's practial approach helps students to understand the idea that reading DNA from a position one base later will give a totally different protein. Through the example of green fluorescent protein students learn how biologists use informatics to help locate new genes of interest which is important in the development of new breeds and varieties.
The concept of Open Reading Frames seems quite complex at first, but this activity with it's practial approach helps students to understand the idea that reading DNA from a position one base later will give a totally different protein. Through the example of green fluorescent protein students learn how biologists use informatics to help locate new genes of interest which is important in the development of new breeds and varieties.
Lesson Description
Guiding Questions
What does the, "one gene one protein" concept tell us about the connections between genes and proteins?
Why must an open reading frame contain sufficient nucleotides to code for a polypeptide chain?
What happens to the protein translated ig you start reading the mRNA one nucleotide further on from the start codon?
Activity 1 - Introduction video from MIT on ORFs
Watch the short video and use the worksheet to make some introductory notes about Open Reading Frames (ORFs)
If a biologist discovers an organism which is green and wants to identify the gene which is responsible for the green protein where do they begin?
It sounds rather confusing but let's imagine that you are looking for the gene which causes "Blobby" to be Green.
Complete the questions on the ![]() ORF Introduction student sheet
ORF Introduction student sheet
Note: If you want to look up the genome of a bacterium called "Buddy" (Yes it really does exist!)
you can do that on the NCBI website Buddy genome page.
Activity 2 - Exploring a genome to find a gene of interest.
The story in activity 1 is very similar to the story of the discovery of the Green Fluorescent Protein in Pacific jelly fish.
Follow the slides and answer the questions below.
Answer these questions about the slides.
Questions
Which of the following techniques of bioinformatics were used in the discovery of the GFP gene:
- Cloning of bacteria to make a cDNA library of the Jellyfish genome.
- Genetic modification of plasmids by adding fragments of the jellyfish genome to them.
- Storage of an organism genome on a database.
- PCR to amplify the DNA in the sample.
- Database tools to search a genome database for an ORF.
- BLAST search to identify the GFP (protein) in other species.
Answer:
- Cloning of bacteria to make a cDNA library of the Jellyfish genome.
- Genetic modification of plasmids by adding fragments of the jellyfish genome to them.
- Storage of an organism genome on a database.
PCR to amplify the DNA in the sample.- Database tools to search a genome database for an ORF.
BLAST search to identify the GFP (protein) in other species.
Activity 3 - Getting practical: finding an ORF yourself
This is a short activity to get you using some of the web tools which help us search and analyse databases of gene and protein data.
Here are the steps;
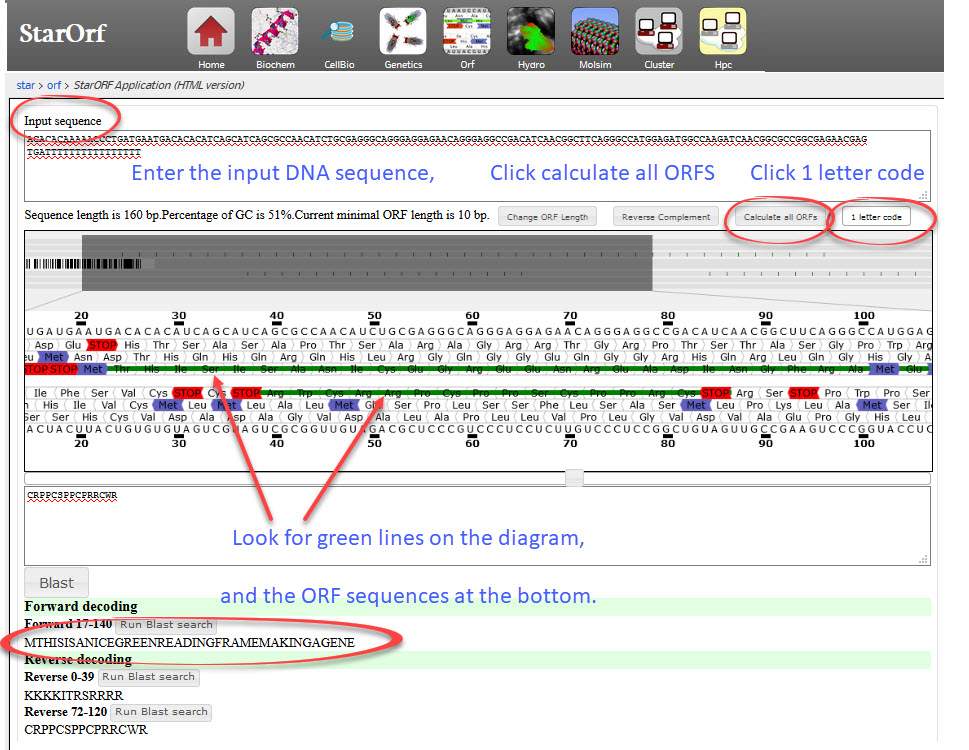
- Write a message with 30 or more letters in Amino Acid single letter code. Remember to avoid letters B, J, O and U
e.g. "THISISANICEGREENREADINGFRAMEMAKINGAGENE"
(It says, "this is a nice green reading frame making a gene") - Convert this to DNA code using EMBOSS Backtranseq
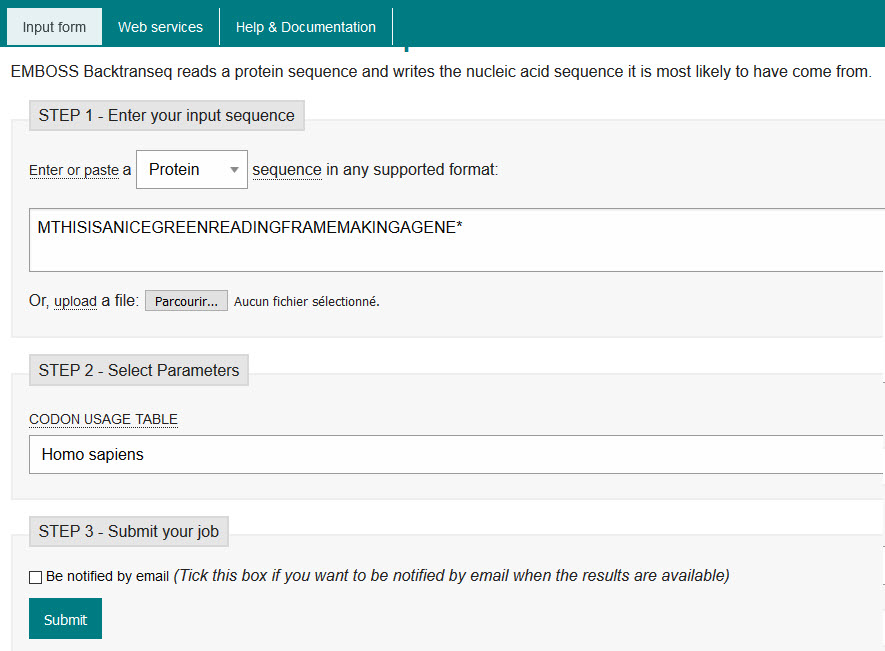
- Add a start codon ATG and a stop codon TGA
- Camoflage this DNA code by adding random DNA codons in front and behind, e.g. AAAAATTTTT
Use this DNA code if you wish:
ACACACAAAAACCCTGATGAATGACACACATCAGCATCAGCGCCAACATCTGCGAGGGCAGGGAGGAGAA
CAGGGAGGCCGACATCAACGGCTTCAGGGCCATGGAGATGGCCAAGATCAACGGCGCCGGCGAGAACGAG TGATTTTTTTTTTTTTTTT - Search for the ORF using the DNA code and this very simple website EMBOSS Sixpack
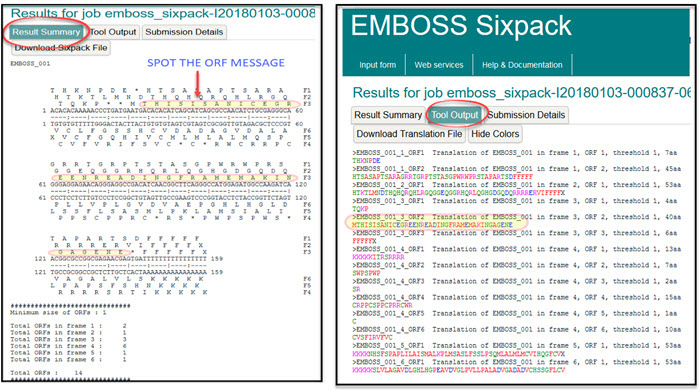
- Try the same search in STAR:Orf
Extension Activity - Find the ORF for green fluorescent protein.
This is the cDNA sequence for the GFP protein ( P42212.1 ), coding for 268 amino acids.
MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTL VTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLV NRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLAD HYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
Try to find the ORF for GFP using STAR:Orf
For an extra challenge try the same thing using NCBI's ORFfinder. This is a very powerful tool, not a visual as StarORF.
Teacher's notes
Activity 1 is a gentle introduction to a topic which can become confusing through the complexity of the tool being introduced. For some students this may be enough.
Activity 2 tries to give the use of ORF in the persuit of genes of interest a real life contect using the discovery of the green fluorescent protein. The methods of creating a cDNA library of the jellyfish genome are not required i nthe IB guide but it is not possible to explain the use of ORF finding tools without reference to cDNA libraries.
Activity 3 is a simple hands on attempt to use some of the tools, using a simple amino acid sequence which can be read as words in the SLC of the protein databases, students will be able to identify the correct ORF easily by reading the words in the SLC letters. This links back to an earlier activity which introduced the SLC.
There is a lot of scope for further reading, although students are not required to know the details of green fluorescent protein, they need simply to understand that an ORF must contain enough DNA code for the length of the protein. They sould also understand the application of ORF finding tools in the persuit of genes of interest.
Further details about green fluorescent protein can be found here.
More details about the 2008 Nobel prize for Chemistry, for work on GFP, can be found on this website.
A nice ORF Activity and explanation from Amrita - more complex than required fror IB but clearly explained.
A really detailed and clear explanations of Identifying, Analysing and Sequencing cloned DNA is possible useful background for teachers.
The original publication by Prascher et al (1992) can be read here in pdf format.
This final video could be useful for interested students or teachers. It goes a little further into the use of databases for find genes.
Activity 4 - Exploring a genome using UGENE and FASTA
This ![]() complex activity on ORFs would be nicely adapted into a simpler activity illustrating the tools
complex activity on ORFs would be nicely adapted into a simpler activity illustrating the tools
BLAST (in a later activity)
Nice NCBI video explaining some of the features of the NCBI BLAST tool

 IB Docs (2) Team
IB Docs (2) Team