Identifying Polysomes
- Translation can occur simultaneously at multiple positions along mRNA
- Polysomes (or polyribosomes) are groups of two or more ribosomes translating the same mRNA transcript
- Multiple copies of the same polypeptide chain can be made simultaneously from a single mRNA transcript
- Polysomes effectively increase the amount of polypeptide produced
- In electron micrographs, polysomes look like ‘beads on a string’ with each bead representing a ribosome
- There are visible differences between eukaryotes and prokaryotes
- In prokaryotes, the lack of a nucleus means transcription and translation are coupled
- Translation starts before the mRNA has finished being transcribed from the DNA
- On an electron micrograph, multiple polysomes can appear on growing mRNA strands along the DNA molecule
- In eukaryotes, mRNA is transported out of the nucleus prior to translation
- On an electron micrograph, polysomes are seen on the mRNA with no involvement of DNA
- As ribosomes move in the same 5’ to 3’ direction along the mRNA, ribosomes towards the 3’ end have longer polypeptide chains being synthesised
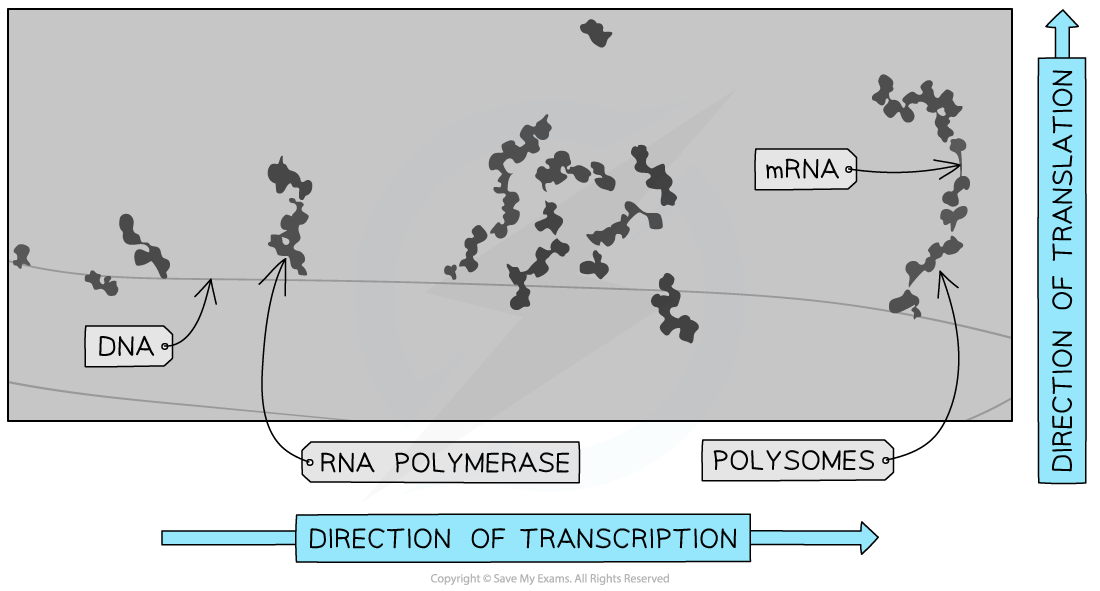
Electron micrograph of prokaryotic polysomes, the image shows simultaneous transcription and translation of a bacterial gene
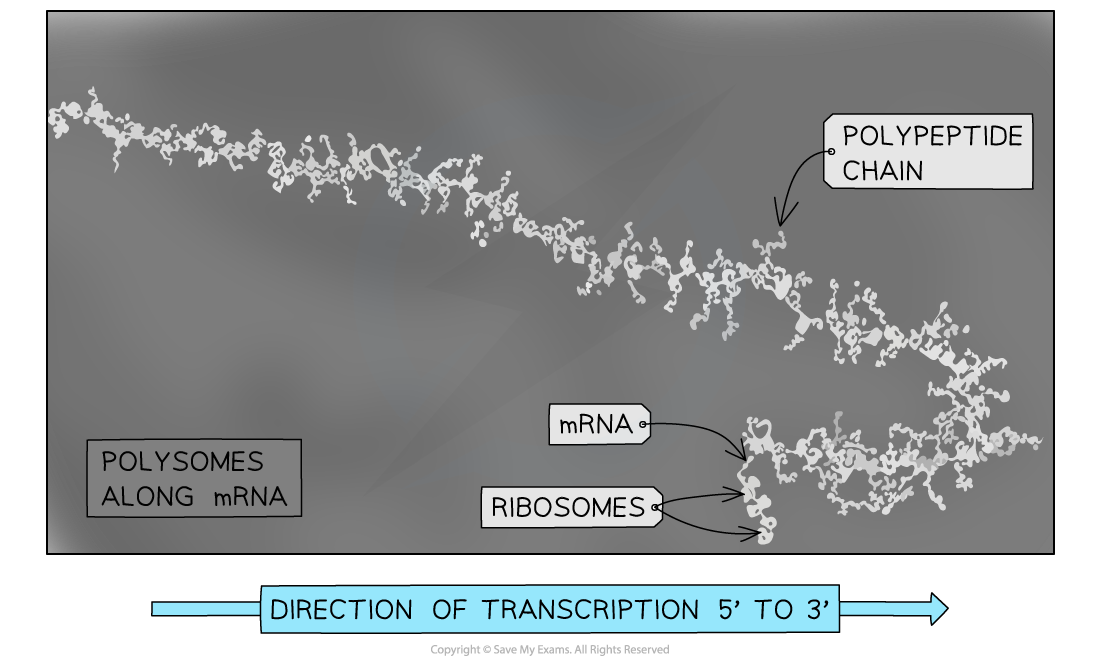
Electron micrograph of eukaryotic polysomes, polypeptide chains can be seen emerging from the ribosomes
Visualising the Structure of Ribosomes
- There are many ‘open source’ databases that contain data relating to the three-dimensional structure of proteins
- The most commonly used ones are PDB (Protein Data Bank) and Swiss-Prot
- Such databases allow researchers to analyse biological molecules and study interactions between them
- This helps relate structure to biological function
- Data relating to the three-dimensional structure of biological molecules can be visualised using molecular visualisation software such as Mol* or Jmol
- Molecules can be represented in many different ways including ball and stick atom models or simplified ribbon representations that show the protein backbone
- Most molecular visualisation software is freely available on the Internet or can be accessed through many bioinformatics repositories
Analysing the structure of the eukaryotic ribosome
- Visit the PDB and search for: Yeast 80S ribosome 4V7R (do not put the search term in quotes)
- Select the “3D view” to view the protein structure in mol*
- Eukaryotic ribosome are complex molecules consisting predominantly of ribosomal RNAs and multiple proteins - these are represented in different colours
- The molecule appears to be made of two distinct halves (subunits)
- Rotate or zoom into the image to visualise the different components
- Try and identify the ribosomal RNA in the two subunits (hover the cursor over)
- An opening (groove) between the subunits should be visible - mRNA passes through here
Analysing the structure of a tRNA molecule
- Search for: 1YFG in the PDB (1YFG is the database identifier for yeast initiator tRNA)
- Select the “3D view” to view the protein structure in mol*
- You should be able to recognise the loop structures
- Consider how this structure is specific to the tRNA-activating enzyme
- Try and identify the anticodon region and amino acid binding site (acceptor arm)
- Try selecting a different viewer such as JSmol
- Investigate changing settings in the viewer
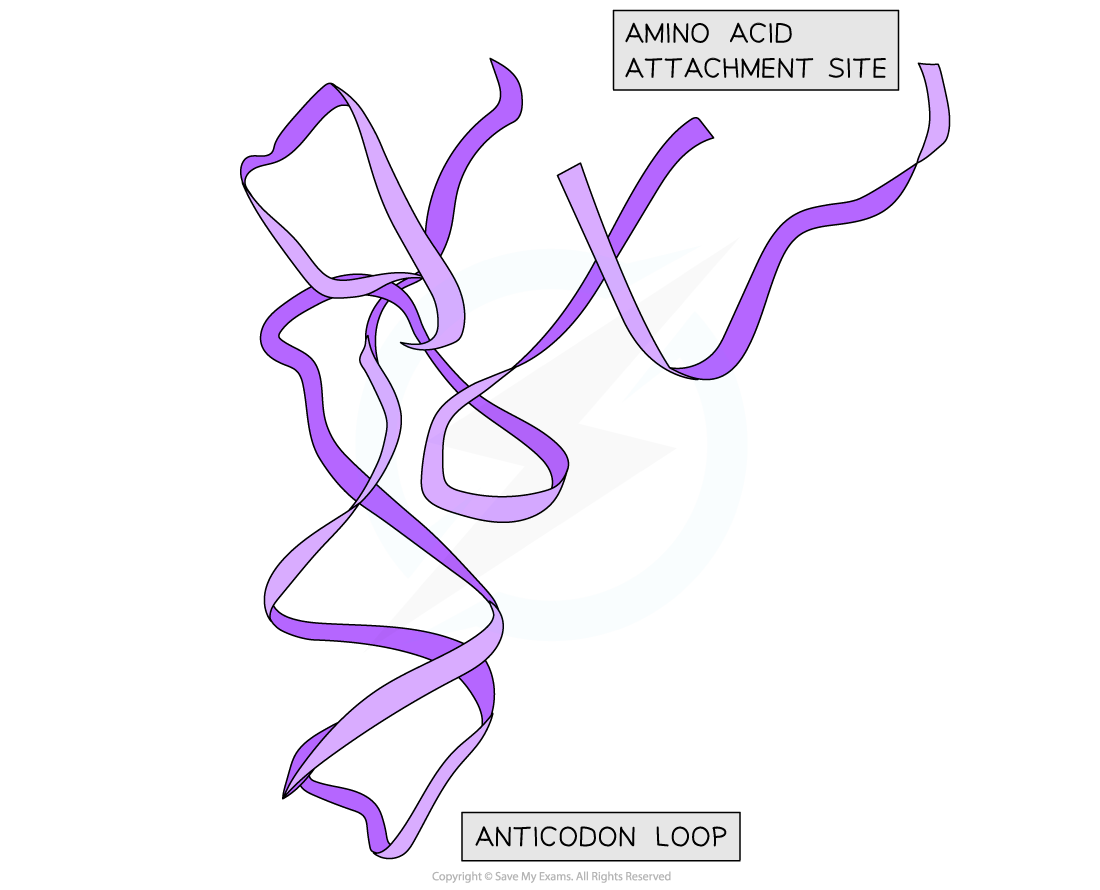
Structure of yeast tRNA
