Initiation of Translation
Initiation of translation involves assembly of the components that carry out the process.
- During translation, the specific sequence of messenger RNA (mRNA) is translated to produce a polypeptide chain consisting of amino acids
- mRNA is a single stranded, linear, RNA molecule that transfers the information in DNA from the nucleus into the cytoplasm
- Translation is categorised into three stages: initiation, elongation and termination
- Translation occurs in the cytoplasm at complex molecules made of protein and RNA called ribosomes
- Ribosomes have a two-subunit (large and small) structure that helps bind mRNA
- Ribosomes have three tRNA binding sites termed “E” (exit), “P” (peptidyl) and “A” (aminoacyl)
- At the A site the mRNA codon joins with the tRNA anticodon
- At the P site the amino acids attached to the tRNA are joined by peptide bonds
- At the E site the tRNA exits the ribosome
- Another key molecule in translation is transfer RNA (tRNA) that decodes mRNA
- tRNA molecules are single stranded RNA molecules that fold to form a clover-shaped structure
- The folded structure is held together by hydrogen bonds between bases at different points on the strand
- tRNA molecules are the shortest of the RNA molecules, being only around 80 nucleotides in length
- There are 20 different types of tRNA molecule, one for each of the amino acids involved in protein synthesis
- tRNA molecules have a region that binds to a specific amino acid as well as a three-nucleotide region called an anticodon that is complementary to the codon on mRNA
- The role of tRNA molecule is to carry a specific amino acid to the ribosome
- tRNA molecules are single stranded RNA molecules that fold to form a clover-shaped structure
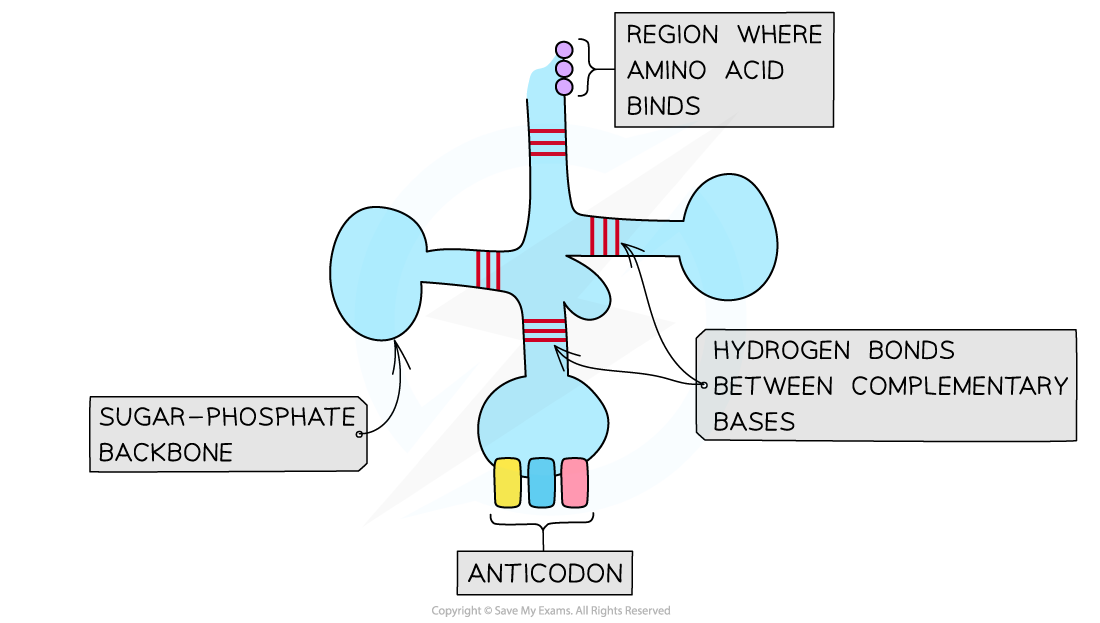
Structure of tRNA
- In eukaryotic cells, the mRNA molecule leaves the nucleus through the nuclear pores
- Translation is initiated by the following process
- A small ribosomal subunit attaches to the 5’ end of mRNA
- An initiator tRNA molecule carrying the amino acid methionine binds to the small ribosomal subunit
- The initiator tRNA occupies the “P” site on the ribosome
- The ribosome moves along the mRNA until it locates a start codon (AUG)
- The large ribosomal subunit binds to the small subunit
- Elongation of the polypeptide can begin
Elongation of the Polypeptide
- The initiator tRNA currently occupies the “P” site, the next codon on the mRNA signals for the corresponding tRNA to bind at the “A” site
- The two amino acids (attached to the tRNAs) are linked with a peptide bond, forming a dipeptide
- Synthesis of the peptide chain now involves a repeated cycle of events
- In the cytoplasm, free tRNA molecules bind to their corresponding amino acids and transport them to the ribosome
- The ribosome shifts along the mRNA one codon (three bases) at a time
- The initiator tRNA in the “P” site moves to the “E” site which releases it
- The tRNA carrying the peptide chain moves from the “A” site to the “P” site
- The next mRNA codon is exposed and a tRNA with the complementary anticodon binds to the unoccupied “A” site whilst its amino acid is linked to the polypeptide chain
- The cyclical process is repeated as new amino acids are added to the growing chain
Termination of Translation
- The process of elongation continues until one of three ‘stop’ codons (UAA, UAG and UGA) on the mRNA molecule is reached
- Stop codons do not code for a tRNA molecule but act as a signal for translation to stop
- The polypeptide chain and mRNA are released from the ribosome
- The ribosome disassembles back into two separate subunits
- And can await the arrival of the next mRNA molecule
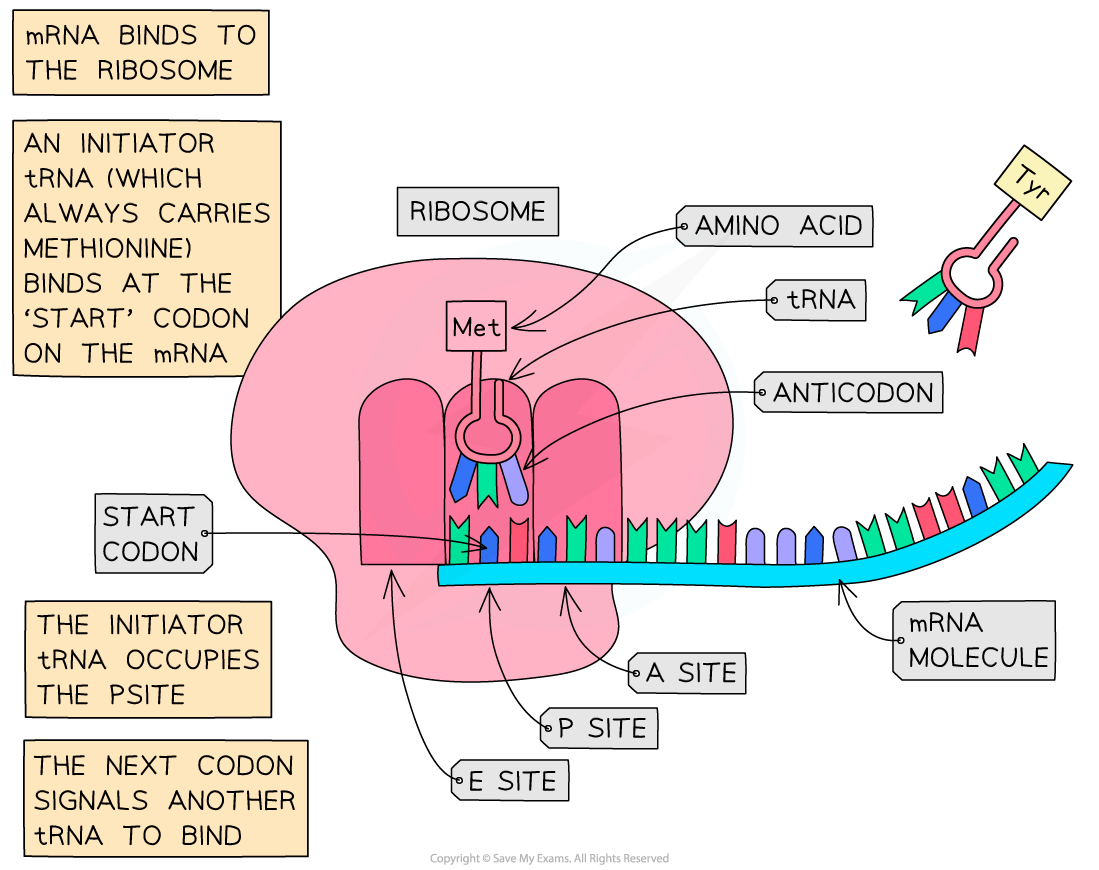
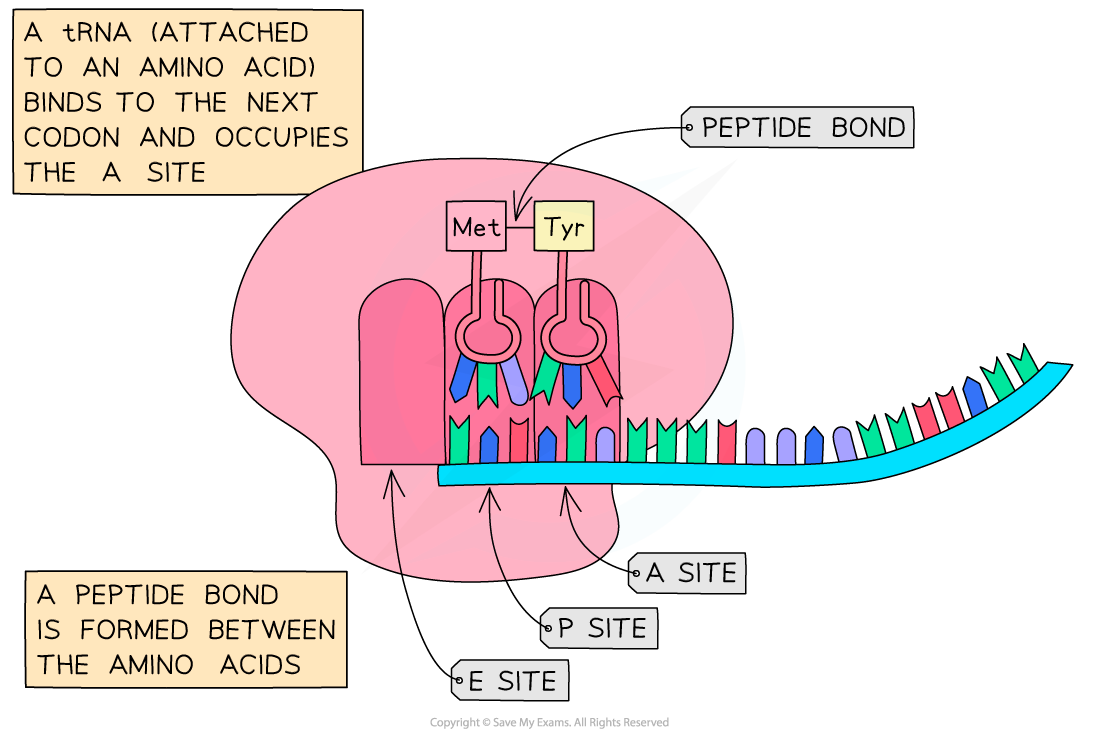
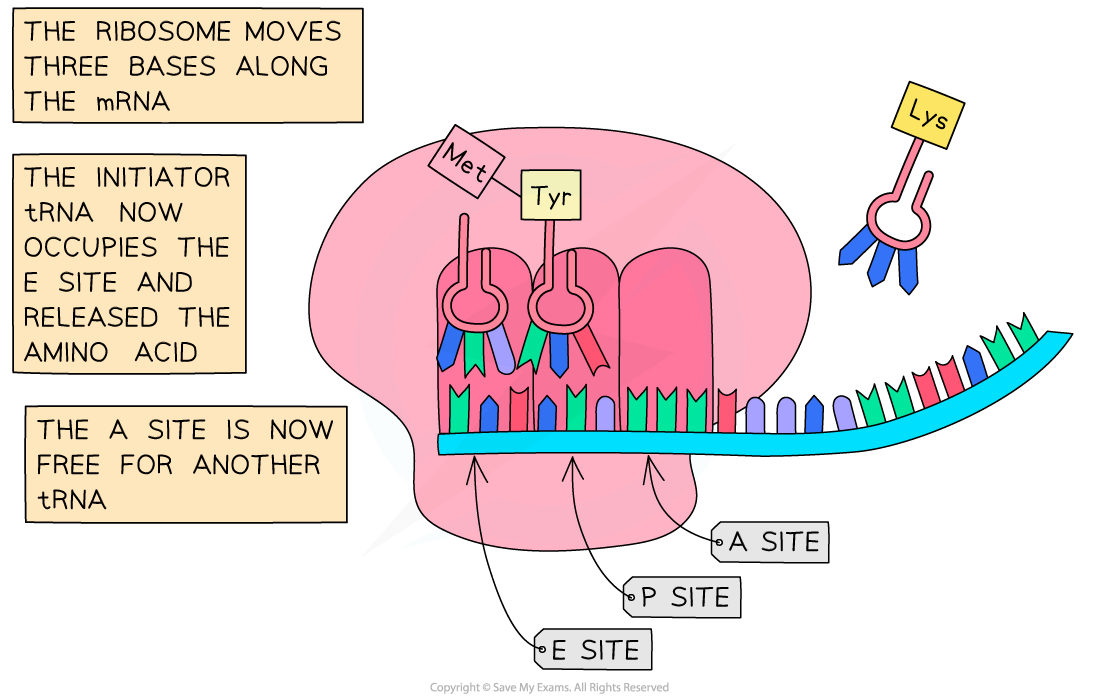
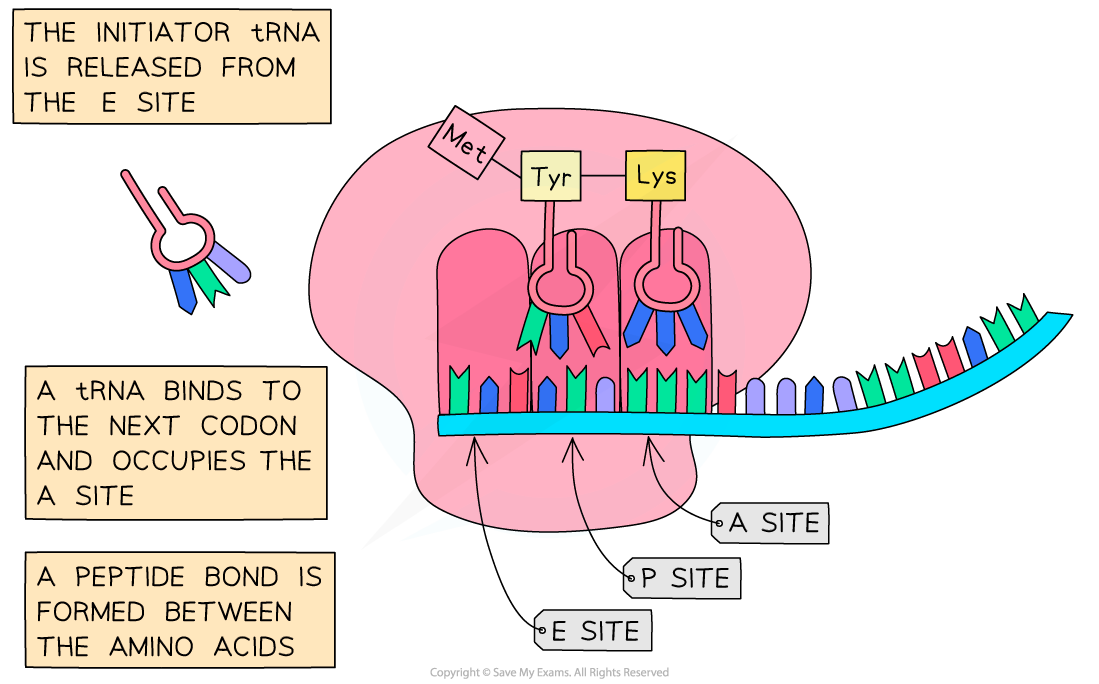
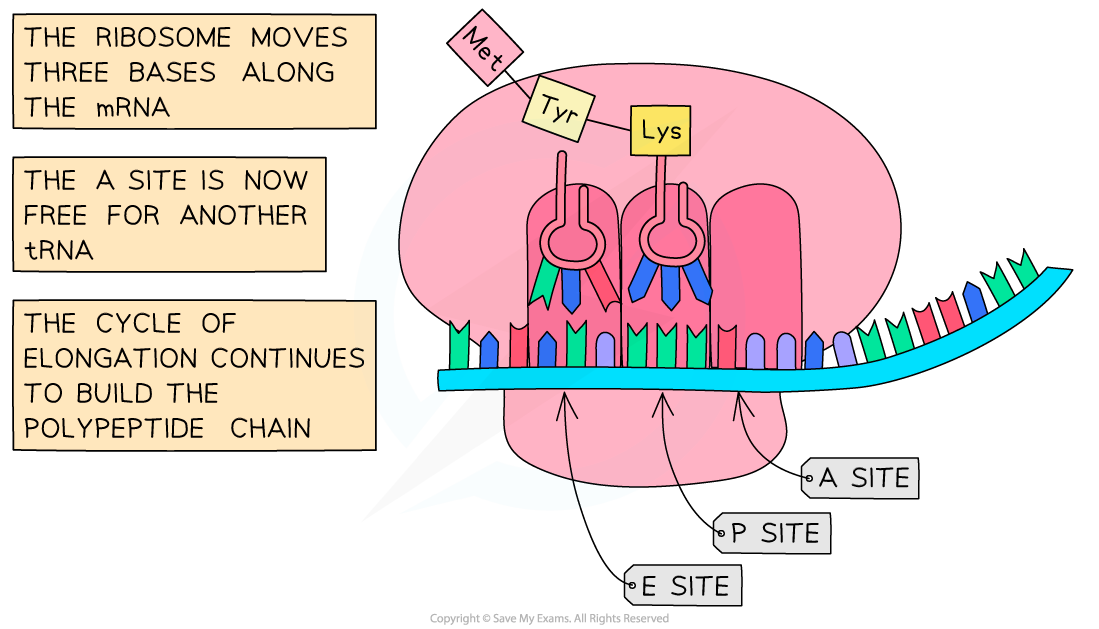
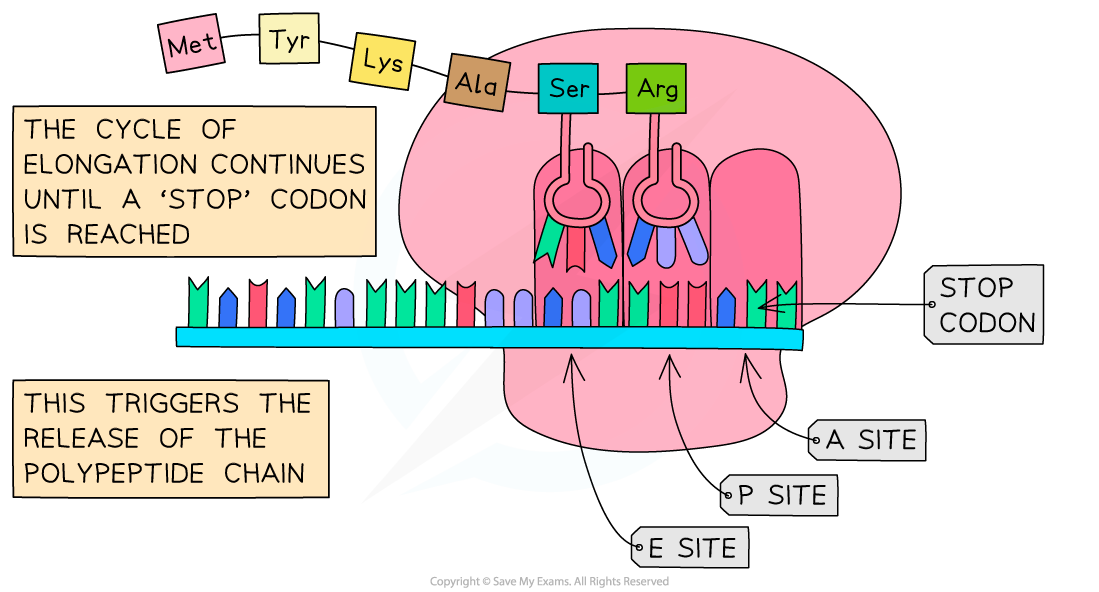
Following the initiation of protein synthesis, translation involves a repeated cycle of events to build the polypeptide chain, tRNA molecules move into the A, P and E sites as the ribosome reads the mRNA
Exam Tip
You don't need to remember the precise base sequences of start and stop codons for your examination.
tRNA-activating Enzymes
- Amino acids are paired to specific tRNA molecules through the action of tRNA-activating enzymes
- Each tRNA activating enzyme recognises a specific tRNA molecule
- tRNA-activating enzymes, in common with most enzymes, are substrate-specific and recognise the correct tRNA molecules by their shape
- Nucleotide sequence variability between tRNA molecules results in variation in their three-dimensional structure
- Active sites of tRNA-activating enzymes are optimised to bind a specific tRNA
- Initially, a tRNA-activating enzyme binds to ATP and a specific amino acid
- The active site of the enzyme attracts a conformationally-specific tRNA molecule
- The tRNA molecule is bound to the amino acid using ATP (phosphorylation) to create a high energy bond
- The stored energy in this bond will be used later in peptide bond formation to link the amino acid to the growing polypeptide chain
- This is an example of how an anabolic reaction like protein synthesis utilises the energy stored in ATP
- A tRNA molecule with an amino acid attached is called a charged tRNA
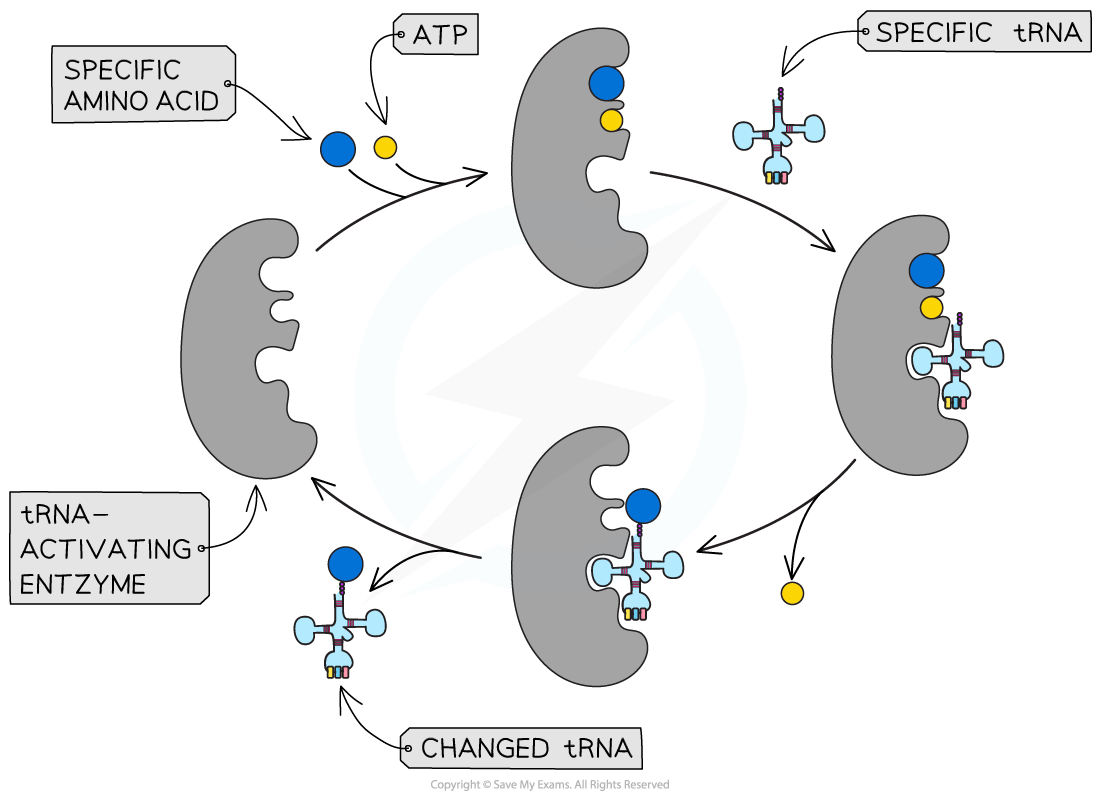
Specific tRNA-activating enzymes are involved in charging an amino acid to a specific tRNA molecule
