Exceptions to Mendel's Rules
NOS: Mendel used observations of the natural world to find and explain patterns and trends
- Since Mendel, scientists have looked for discrepancies and asked questions based on further observations to show exceptions to the rules. For example, Morgan discovered non-Mendelian ratios in his experiments with Drosophila
- When looking at dihybrid crosses (crosses with two pairs of observable characteristics), Mendel explained his experimental data with his law of independent assortment
- That individual characteristics are inherited completely independently of each other
- In many cases, this is correct
- The significance of Mendel's work went largely unnoticed for decades, until after his death in 1884
- Scientists in the 1890s and early 1900s picked up his experimental findings and replicated them
- The large number of trials that Mendel undertook highlighted patterns/trends in the inheritance of certain factors (what are now known as genes)
- However, discrepancies were noticed when scientists replicated Mendel's experiments
- And also when experiments were undertaken on the inheritance of certain genes in other organisms
- Many of their dihybrid crosses replicated the 9:3:3:1 pattern of phenotypes that Mendel first observed in his work
- William Bateson and Reginald Punnett, two Cambridge University geneticists (in collaboration with a third Cambridge biologist, Edith Saunders), replicated Mendel's findings, again in experiments with sweet peas, however
- They discovered some apparently anomalous results in certain cases, in which phenotype ratios did not follow the classical 9:3:3:1 pattern
- Many scientists would have dismissed non-conforming results as mere anomalies, however Bateson, Punnett and Saunders chose to search for an explanation
- Punnett's quote from one his laboratory notebooks sums up their approach:
- "Treasure your exceptions! When there are none, the work gets so dull that no one cares to carry it further."
- Bateson and Punnett performed further work, mainly on crossings of sweet peas and crossings of chickens, but were unable to offer a robust explanation for certain unpredictable phenotype ratios in their crosses
Gene Linkage
Gene loci are said to be linked if they are on the same chromosome
- Loci (singular: locus) refers to the specific linear positions on the chromosome that genes occupy
- If genes are on the sex chromosome, they are said to be sex-linked
- Sex-linked genes have characteristics that generally only affect one gender of a species
- These genes are usually on the X chromosome because the Y chromosome contains very few genes
- In humans, colour-blindness and haemophilia are notable examples of genetic conditions that only affect males
- Linked genes located on the chromosomes 1-22, or any chromosome that is not a sex chromosome (called autosomes) are said to be examples of autosomal linkage
- The likelihood of genes being inherited together, or the extent to which they are linked, is measured in units called centimorgans, in honour of Thomas Hunt Morgan's work
Notation for link genes
- When writing linked genotypes it can be easier to keep the linked alleles within a bracket
- For example, an individual has the genotype FFGG. However, if there is linkage between the two genes, it would be written as (FG)(FG)
- Another commonly-used way of denoting linked alleles is to link them with a line. So, for example, linkage between genes F and G might be shown as
F G
_____
_____
f g
- Remember to distinguish between sex linkage and autosomal linkage. The explanation of non-Mendelian ratios falls into the domain of autosomal linkage for IB
Autosomal linkage
- Dihybrid crosses and their predictions rely on the assumption that the genes being investigated behave independently of one another during meiosis
- However, not all genes assort independently during meiosis
- Some genes which are located on the same chromosome display autosomal linkage and stay together in the original parental combination
- Linkage between genes affects how parental alleles are passed onto offspring through the gametes
Identifying autosomal linkage from phenotypic ratios
- In the following theoretical example, a dihybrid cross is used to predict the inheritance of two different characteristics in a species of newt
- The genes are for tail length and scale colour
- The gene for tail length has two alleles:
- Dominant allele T produces a normal length tail
- Recessive allele t produces a shorter length tail
- The gene for scale colour has two alleles:
- Dominant allele G produces green scales
- Recessive allele g produces white scales
Without linkage
- The outcomes for this dihybrid cross if the genes are unlinked are as follows
Dihybrid Cross without Linkage Punnett Square Table
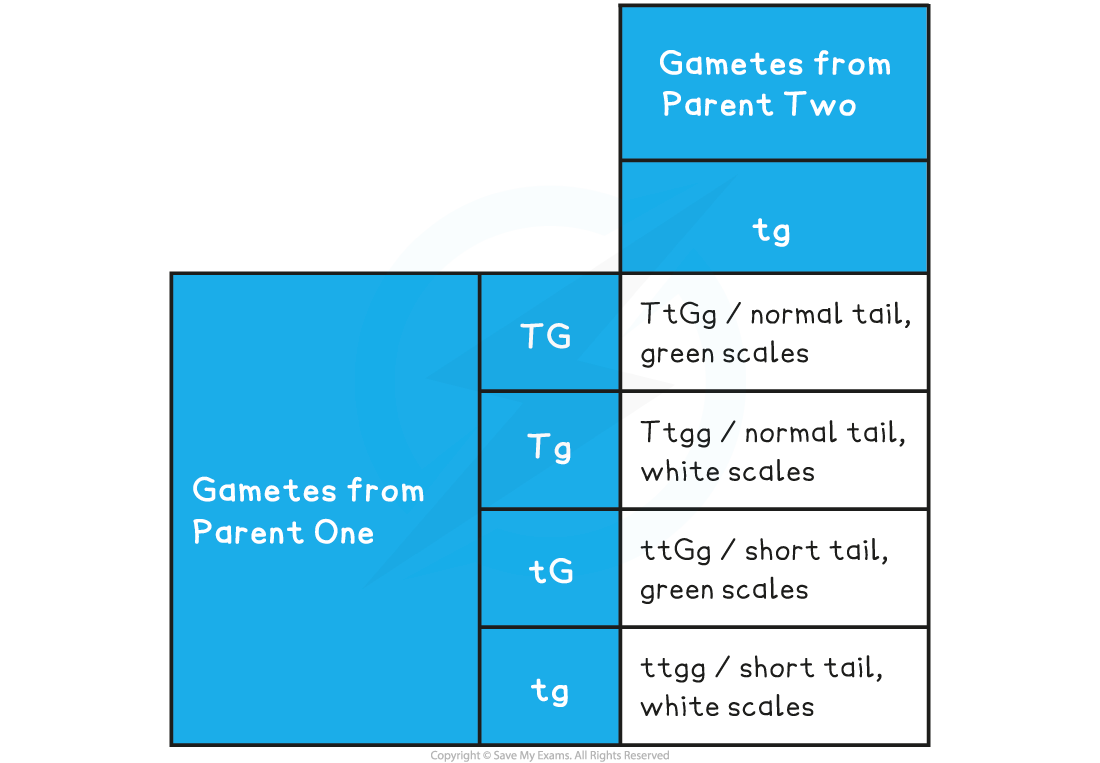
- Predicted ratio of phenotypes in offspring =
- 1 normal tail, green scales : 1 normal tail, white scales : 1 short tail, green scales : 1 short tail, white scales
- Predicted ratio of genotypes in offspring =
- 1 TtGg : 1 Ttgg : 1 ttGg : 1 ttgg
With linkage
- However, if the same dihybrid cross is carried out but this time the genes are linked, we get a different phenotypic ratio
- There would be a 1 : 1 phenotypic ratio (1 normal tail, green scales : 1 short tail, white scales)
- This change in the phenotypic ratio occurs because the genes are located on the same chromosome
- The unexpected phenotypic ratio, therefore, shows us that the genes are linked
- The explanation for this new phenotypic ratio is given in the worked example below:
Worked Example
Worked example: Explaining autosomal linkage
- In reality, the genes for tail length and scale colour in this particular species of newt show autosomal linkage
Parental phenotypes: normal tail, green scales x short tail, white scales
Parental genotypes: (TG)(tg) (tg)(tg)
Parental gametes: (TG) or (tg) (tg)
Dihybrid Cross with Linkage Punnett Square Table
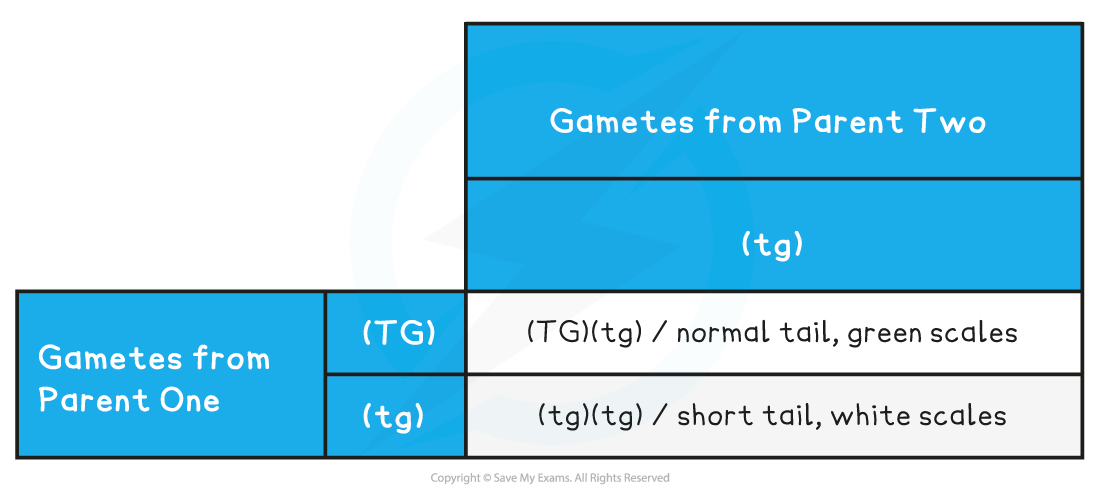
- Predicted ratio of genotypes in offspring =
- 1 (TG)(tg) : 1 (tg)(tg)
- Predicted ratio of phenotypes in offspring =
- 1 normal tail, green scales : 1 short tail, white scales
Exam Tip
When you are working through different genetics questions you may notice that test crosses involving autosomal linkage predict solely parental type offspring (offspring that have the same combination of characteristics as their parents). However in reality recombinant offspring (offspring that have a different combination of characteristics to their parents) are often produced. This is due to the crossing over that occurs during meiosis. The crossing over and exchanging of genetic material breaks the linkage between the genes and recombines the characteristics of the parents. So if a question comes along that asks you why recombinant offspring are present you now know why!
Non-Mendelian Ratios in Drosophila
- In the way that Bateson, Punnett and Saunders developed and refined Mendel's findings, Thomas Hunt Morgan further refined genetic theory
- His work was awarded the Nobel Prize in 1933
A fruit fly (Drosophila melanogaster) with some of the phenotypic variations observed in Thomas Hunt Morgan's work
Sex linkage
- Working in the USA in the early 20th century, he bred fruit flies (Drosophila melanogaster) over successive generations
- In his cross-breeding experiments he came across red-eyed wild types and white-eyed mutants
- He realised there was a distinct sex bias in phenotypic distribution
- All-female offspring of a red-eyed male were red-eyed while all male offspring of a white-eyed female were also white-eyed
- Morgan hypothesised that this occurred because the gene for eye colour was located on a sex chromosome (i.e. X-linked)

Sex linkage in Drosophila. A cross between a homozygous white-eyed female and a male with red eyes gives all white-eyed males and red-eyed female offspring
Autosomal linkage
- As Morgan continued his experiments he noticed a number of different traits in fruit flies that did not conform to Mendelian ratios as several phenotypes occurred in much lower frequencies than expected
- Based on this data, Morgan made two key proposals:
- The alleles for these traits were located on the same chromosome (gene linkage) meaning they did not independently assort
- Linked alleles could be unlinked via recombination (crossing over) to produce recombinant offspring (offspring that have a different combination of characteristics to their parents)
- This is due to the crossing over that occurs during meiosis
- The crossing over and exchanging of genetic material breaks the linkage between the genes and recombines the characteristics of the parents
- Morgan also observed that the number of recombinants that resulted from crossing linked genes varied depending on the combination of traits
- He proposed the idea that the number of recombinants (crossover frequency) may be related to the distance between two genes on the same chromosome
- Genes that were further apart had a higher crossover frequency, whereas genes closer together exhibited a lower crossover frequency
- Morgan used this concept to create the first gene linkage maps
- These maps displayed the relative positions of genes on chromosomes
