Dichotomous Keys
- For anyone who doesn't specialise in studying a particular group of organisms, it can be very difficult to identify, or ID, a species when working in the field
- Correctly identifying species may be important for a researcher studying biodiversity or looking at the impacts of a changing environment on a community of organisms
- Someone seeking to identify species with which they are not already familiar may use a tool known as a dichotomous key
- A dichotomous key contains a series of paired statements
- The term 'dichotomous' refers to these pairs of statements
- An example of such a pair of statements might read:
- The organism shows radial symmetry
- The organism shows bilateral symmetry
- Or:
- The organism has one pair of wings
- The organism has two pairs of wings
- To work through a dichotomous key, you start with the first pair of statements and apply them to the unknown species; one statement will be clearly false, while the other will be a correct description of the species
- The correct statement leads to another pair of statements, and so on until the final correct statement leads to the name of the species
Worked Example
Use the dichotomous key provided to identify the type of organism below
The organism has an exoskeleton, a segmented body, and 4 pairs of legs. It has no tail and cannot produce silk
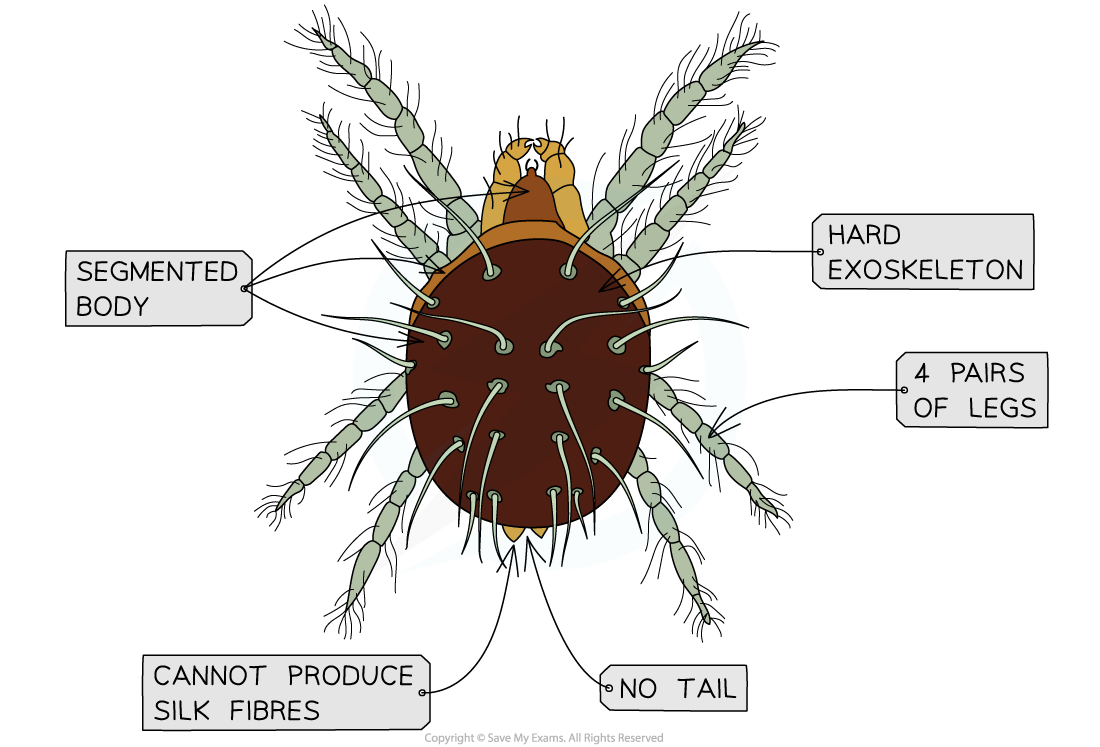

Step 1: Decide which of the first pair of statements applies
The organism has an exoskeleton, so we can ignore the first statement in pair 1 and move onto the third pair of statements as instructed in the key
Step 2: Decide which of the third pair of statements applies
The organism has 4 pairs of legs, so we move on to the fourth pair of statements as instructed
Step 3: Decide which of the fourth pair of statements applies
The organism has no tail, so we know that it is not a scorpion, and we move onto the sixth pair of statements as instructed
Step 4: Decide which of the sixth pair of statements applies
The organism cannot produce silk, so it must be a species of mite
Constructing a dichotomous key
- When constructing a key, the following should be considered
- Each pair of statements should contain features that are clearly identifiable and not subject to opinion e.g. the organism has 3 pairs of legs, rather than e.g. the organism is small
- A leg count gives an objective number, but size is relative and depends on what the species is being compared to
- Statements must be 'yes' or 'no' in style
- Each pair of statements should divide the organisms being identified into two distinct groups
- Each subsequent pair of statements should divide the organisms into smaller and smaller groups
- Each statement should be followed by either a number to continue the process of narrowing down the options, or should name the organism to which it applies
- Each pair of statements should contain features that are clearly identifiable and not subject to opinion e.g. the organism has 3 pairs of legs, rather than e.g. the organism is small
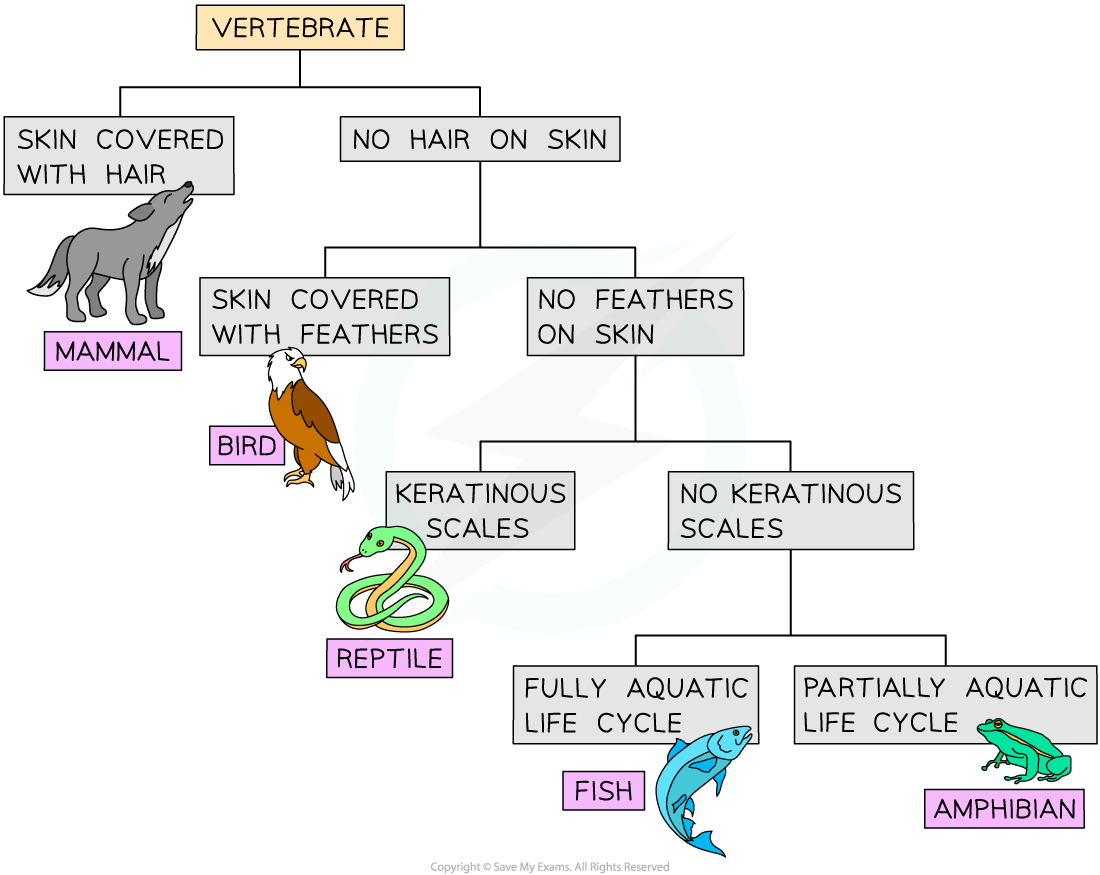
Dichotomous keys can also be represented using a branched diagram as shown here. This visual representation can be easier to understand, but it is limited in the number of organisms it can include
Exam Tip
Get some practice at using a dichotomous key by identifying the remaining organisms featured in the key, all of which are described below. Note: they are all quite easily recognisable animals so you should be able to tell whether you have used the key correctly or not.
- The organism has an exoskeleton, a segmented body, and 4 pairs of legs. It has a tail with a sting
- The organism has an exoskeleton, a segmented body, and 3 pairs of legs. Its body is uniform in colour
- The organism has an exoskeleton, a segmented body, and 3 pairs of legs. It's body is striped black and yellow
- The organism is soft bodied with a muscular foot and a hard shell
- The organism has an exoskeleton, a segmented body, and 4 pairs of legs. It has no tail but can produce silk fibres from a specialised organ on its abdomen
- The organism is soft bodied with a muscular foot and no hard shell
Analysis of Cladograms
- Evolutionary relationships between species can be represented visually using a diagram called a cladogram
- Cladograms are evolutionary trees that show probable order of divergence from ancestral species and therefore probable relationships between species
- Analysis of a cladogram can provide several pieces of information
- The point at which two branches separate is known as a node, and represents common ancestor species
- A node immediately adjacent to a pair of clades indicates that these two clades share a recent common ancestor
- This shows that the two clades are more closely related to each other than they are to any other clade in the cladogram
- If several nodes need to be traced back before two clades can be joined, this indicates a more distant relationship between two clades
- Cladograms sometimes show numbers along the branches; these indicate the number of base or amino acid changes that have occurred between one node and the next or between a node and an emerging clade or species
- The constant rate at which mutations accumulate means that these numbers can be used as a molecular clock to calculate how much time has passed
- The point at which two branches separate is known as a node, and represents common ancestor species
- Some cladograms have a time scale to show how many millions have years have passed
- This is done using the principle of parsimony which states that the simplest explanation is preferred
- The computer builds the shortest possible cladogram with the smallest number of divergence events to fit the available data
- The reliability of a cladogram may vary depending on the amount of sequence data used to construct it
- A cladogram based on the sequencing of one gene will be less reliable than a cladogram based on the sequencing of several genes
- Cladograms are subject to change when new sequence data becomes available
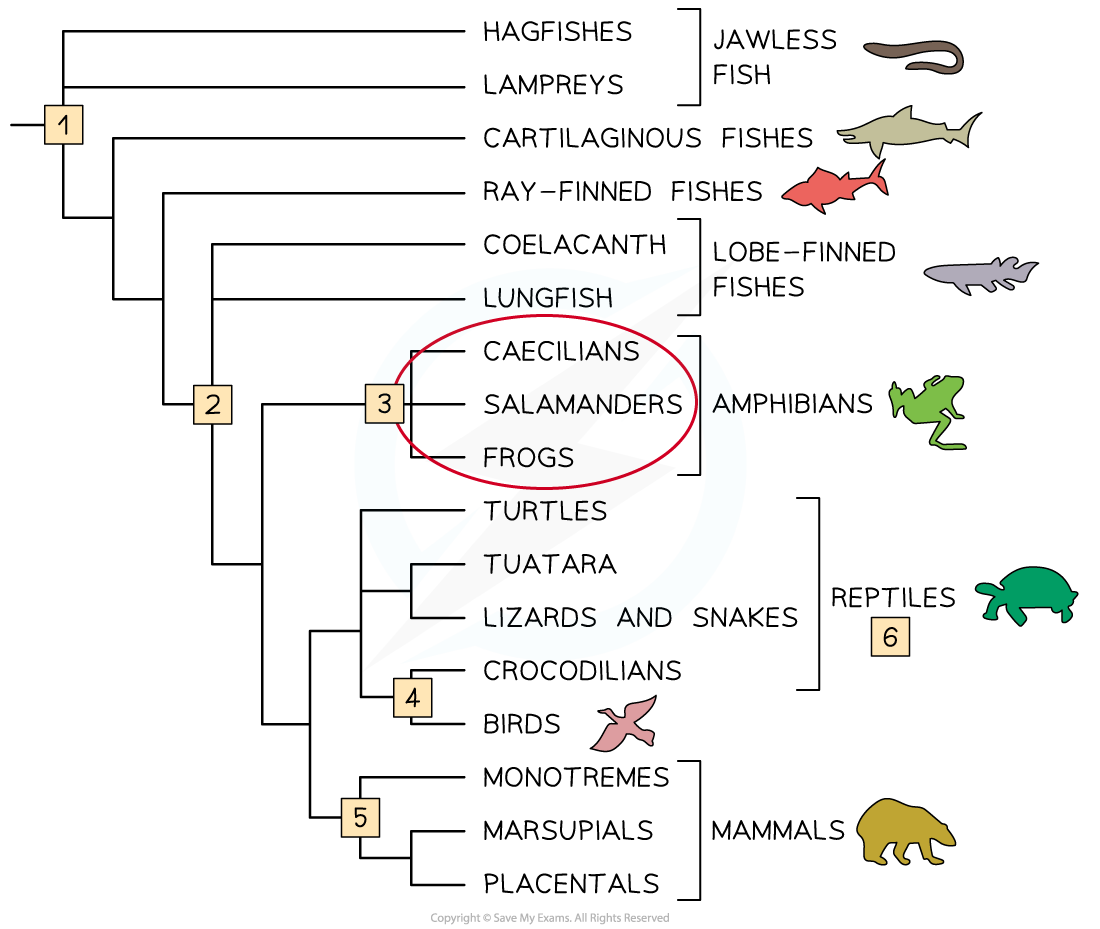

A cladogram with notes to indicate some of the conclusions that can be drawn from it. Note that this cladogram contains no numbers or time scale, so it does not show the number of base or amino acid changes that have occurred between one node and the next, or how much time has passed between nodes.
